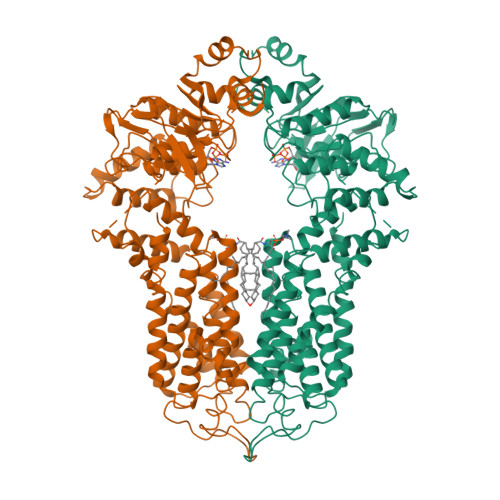
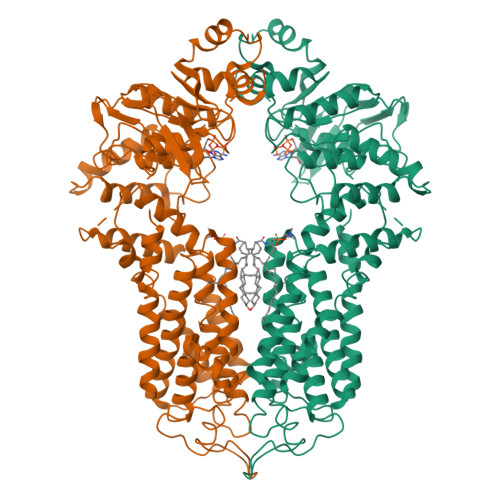
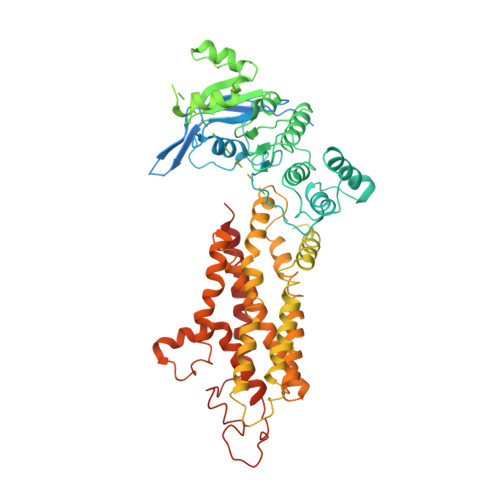
Structure and transport mechanism of the human cholesterol transporter ABCG1.
Xu, D., Li, Y., Yang, F., Sun, C.R., Pan, J., Wang, L., Chen, Z.P., Fang, S.C., Yao, X., Hou, W.T., Zhou, C.Z., Chen, Y.(2022) Cell Rep 38: 110298-110298
- PubMed: 35081353
- DOI: https://doi.org/10.1016/j.celrep.2022.110298
- Primary Citation of Related Structures:
7FDV - PubMed Abstract:
The reverse cholesterol transport pathway is responsible for the maintenance of human cholesterol homeostasis, an imbalance of which usually leads to atherosclerosis. As a key component of this pathway, the ATP-binding cassette transporter ABCG1 forwards cellular cholesterol to the extracellular acceptor nascent high-density lipoprotein (HDL). Here, we report a 3.26-Å cryo-electron microscopy structure of cholesterol-bound ABCG1 in an inward-facing conformation, which represents a turnover condition upon ATP binding. Structural analyses combined with functional assays reveals that a cluster of conserved hydrophobic residues, in addition to two sphingomyelins, constitute a well-defined cholesterol-binding cavity. The exit of this cavity is closed by three pairs of conserved Phe residues, which constitute a hydrophobic path for the release of cholesterol in an acceptor concentration-dependent manner. Overall, we propose an ABCG1-driven cholesterol transport cycle initiated by sphingomyelin-assisted cholesterol recruitment and accomplished by the release of cholesterol to HDL.
Organizational Affiliation:
Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, China.